SE37:/DS5
Sample Set Information
| ID | SE37 |
|---|---|
| Title | Medicago truncatula shoot metabolite analysis using stable isotopes |
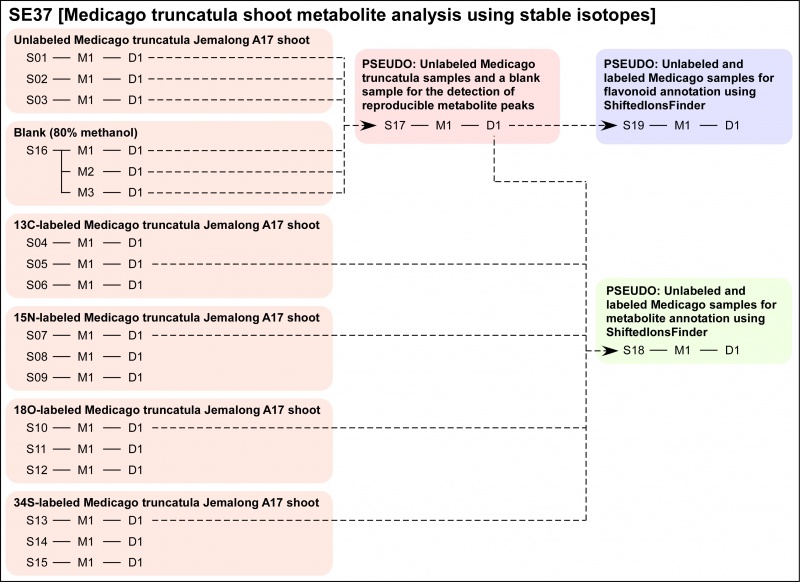
| Description | Metabolites in the Medicago truncatula shoot are investigated by using stable isotope labelings.
This analysis was performed to estimate the elemental composition with high reliability using the number of stable isotopes incorporated in the compound as an index and to add information to discuss the biosynthetic pathway. The Medicago seedlings were labeled with 13C, 15N, 18O, and 34S, respectively in the same condition. Following peak picking by using PowerFT (S01 to S16), the metabolite list was prepared from unlabeled data and blank data (S17). The isotopic peaks were searched and listed using ShiftedIonsFinder (S18). Moreover, the flavonoid-like peaks were also searched using ShiftedIonsFinder (S19). |
| Authors | AUTHERS; Kota Kera1, Yoshiyuki Ogata2, Takeshi Ara1, Yoshiki Nagashima1, Norimoto Shimada1,3, Nozomu Sakurai1. Daisuke Shibata1 and Hideyuki Suzuki1*
AFFILIATION; 1 Kazusa DNA Research Institute, Kisarazu, Chiba 292-0818, Japan; 2 Graduate School of Life and Environmental Sciences, Osaka Prefecture University, Osaka 599-8531, Japan; 3 Present Address, Tokiwa Phytochemical Co.,Ltd. 158 Kinoko, Sakura, Chiba 285-0801, Japan |
| Reference | ShiftedIonsFinder: A standalone Java tool for finding peaks with specified mass differences by comparing the mass spectra of isotope-labeled and unlabeled data sets. Plant Biotechnology (2014) 3: 269-274
Pathway-specific metabolome analysis with 18O2-labeled Medicago truncatula via a mass spectrometry-based approach. Metabolomics (2018) 14:71 |
| Comment |
Data Analysis Details Information
| ID | DS5 |
|---|---|
| Title | Flavonoid like peak search by ShiftedIonsFinder |
| Description | The candidate peaks with chemical modification such as glycosylation and acylation were searched by comparing the peak list having mass information of aglycon of typical flavonoid against the peak list from an unlabeled sample using ShiftedIonsFinder.
The prepared basic sample and shifted samples were imported to ShiftedIonsFinder and the labeled ions were searched with parameters (Max fold: Glc = 3, Rha = 3, GlcUA = 3, Malonyl = 3, Succinyl = 3; Mass difference = 3ppm, RT difference = 60). Flavonoid list (manually prepared) was used for the analysis. The exported file from ShiftedIonsFinder was arranged by Excel. |
| Comment_of_details |