SE37:/MS1
Sample Set Information
| ID | SE37 |
|---|---|
| Title | Medicago truncatula shoot metabolite analysis using stable isotopes |
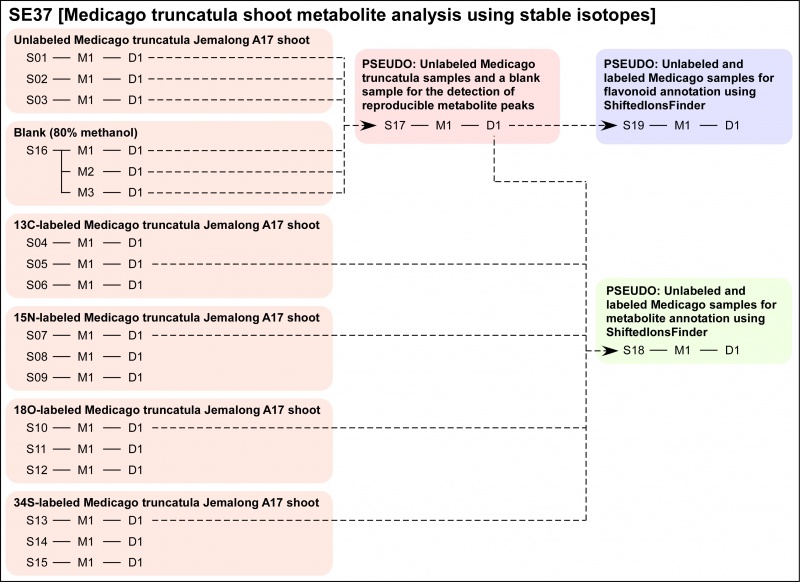
| Description | Metabolites in the Medicago truncatula shoot are investigated by using stable isotope labelings.
This analysis was performed to estimate the elemental composition with high reliability using the number of stable isotopes incorporated in the compound as an index and to add information to discuss the biosynthetic pathway. The Medicago seedlings were labeled with 13C, 15N, 18O, and 34S, respectively in the same condition. Following peak picking by using PowerFT (S01 to S16), the metabolite list was prepared from unlabeled data and blank data (S17). The isotopic peaks were searched and listed using ShiftedIonsFinder (S18). Moreover, the flavonoid-like peaks were also searched using ShiftedIonsFinder (S19). |
| Authors | AUTHERS; Kota Kera1, Yoshiyuki Ogata2, Takeshi Ara1, Yoshiki Nagashima1, Norimoto Shimada1,3, Nozomu Sakurai1. Daisuke Shibata1 and Hideyuki Suzuki1*
AFFILIATION; 1 Kazusa DNA Research Institute, Kisarazu, Chiba 292-0818, Japan; 2 Graduate School of Life and Environmental Sciences, Osaka Prefecture University, Osaka 599-8531, Japan; 3 Present Address, Tokiwa Phytochemical Co.,Ltd. 158 Kinoko, Sakura, Chiba 285-0801, Japan |
| Reference | ShiftedIonsFinder: A standalone Java tool for finding peaks with specified mass differences by comparing the mass spectra of isotope-labeled and unlabeled data sets. Plant Biotechnology (2014) 3: 269-274
Pathway-specific metabolome analysis with 18O2-labeled Medicago truncatula via a mass spectrometry-based approach. Metabolomics (2018) 14:71 |
| Comment |
Analytical Method Details Information
| ID | MS1 |
|---|---|
| Title | Metabolites extraction with 80% methanol and analysis by LC-Orbitrap-MS |
| Instrument | Agilent1100 HPLC (Agilent), LTQ-Orbitrap (Thermo Fisher Scientific) |
| Instrument Type | LC-Orbitrap-MS |
| Ionization | ESI |
| Ion Mode | Positive |
| Description | <Blank preparation>
80% methanol was prepared from methanol (Optima LC/MS, Fisher Chemical) and water (Optima LC/MS, Fisher Chemical) and cleaned up with a disc filter (millex-LG, 0.20um, MILLIPORE).
The metabolites were extracted with 80% methanol (see "Blank preparation") as follows: 1) Measure about 10mg of plant sample in 1.5ml microtube. 2) Ground the sample in the tube with pestle. 3) Add 20 times the volume of cool 80% methanol and mixed well. 4) Incubate it on ice for 30min and then centrifuge it at 15,000/g for 20 min at 4 degree. 5) Collect the supernatants in a new glass vial. 6) Repeat steps 3 to 5 once. 7) Combine all the supernatants in same glass vial. 8) Clean up the solution with the disc filter (millex-LG, 0.20um, MILLIPORE).
A 10ul sample is injected into HPLC. HPLC conditions: Agilent 1100 series (Agilent), Column: TSKgel ODS-100V (4.6 x 250mm, 5 micrometer; TOSOH), Solvent: A; 0.1% formic acid aq. B; ACN (addition 0.1% formic acid fc.), Gradient: (B);3 to 97% (0.0 to 45.0min), 97% (45.1 to 50.0min), 3% (50.1 to 57.0min), Column temp.: 40 degree C, Flow rate=0.5mL/min, PDA: 190-950nm (2nm step). Orbitrap-MS conditions: Filter 1: FTMS + c norm !corona !pi res=60000 o(100.0-1500.0); 2: ITMS + c norm !corona !pi Dep MS/MS Most intense ion from (1); 3: ITMS + c norm !corona !pi Dep MS/MS 2nd most intense ion from (1); 4: ITMS + c norm !corona !pi Dep MS/MS 3rd most intense ion from (1); 5: ITMS + c norm !corona !pi Dep MS/MS 4th most intense ion from (1); 6: ITMS + c norm !corona !pi Dep MS/MS 5th most intense ion from (1)., Rejected mass=235.00000, 376.00000, 609.00000, 810.00000, 1123.00000. |
| Comment_of_details |